mirror of
https://github.com/Bubberstation/Bubberstation.git
synced 2026-01-15 19:46:38 +00:00
## About The Pull Request Did you know that there's 2 types of disks Science can print? One of them creates a whole new techweb and is used to transfer data to and from techwebs (which used to be for Theft objective, but nowadays all it's used for is Admin techweb). The other one is these disks you can find from Lavaland & for the Limbgrower which holds nodes on them directly, they don't need anything else. These are both printable and have very similar names which could easily confuse people, especially since they are both printed and used at the same place, being the R&D room and R&D console. This will hopefully simplify it, by removing the base Tech disks from being printable. The only one that can be printed now is for Limbgrowers, which can't be easily mixed with the other type of Disk. Outside of that, Lavaland disks are staying the same, but I've moved Bepis disks to use this, which allows us to remove Bepis techwebs being made every single time a new bepis disk is created. Examples of it in-game 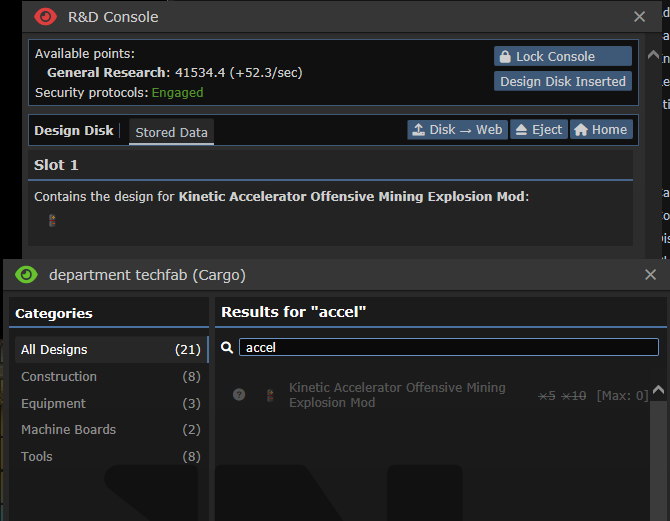 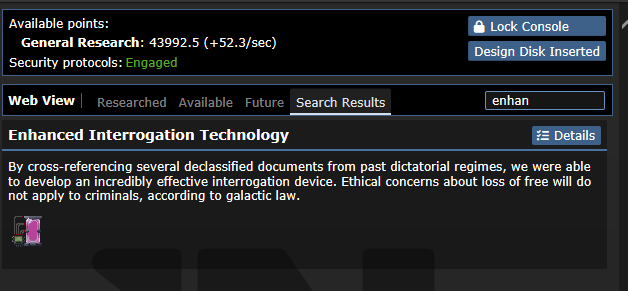 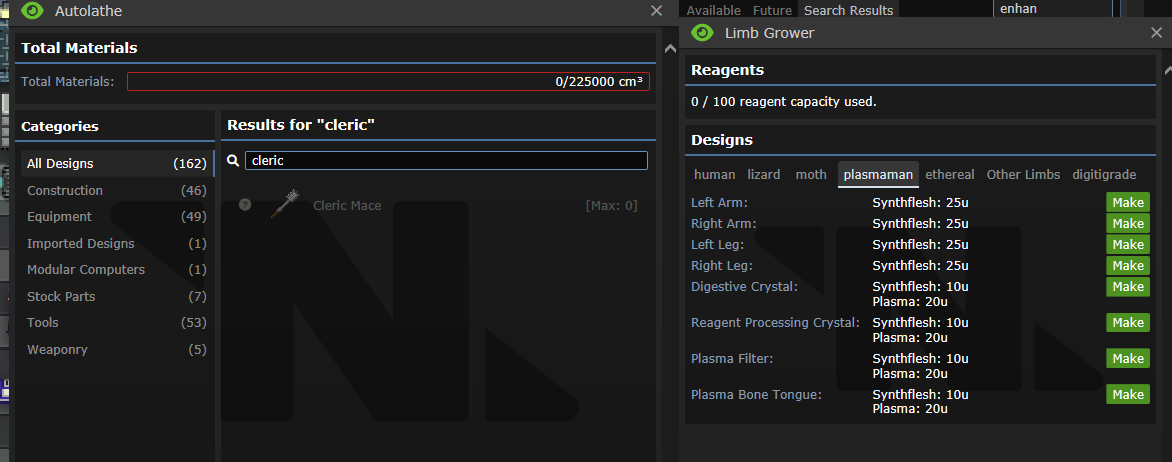 ## Why It's Good For The Game There's no reason why every single Bepis node should be making a whole new techweb set with experiments to complete, roundstart nodes researched, being constantly checked if it should gain research points, have a list of papers to publish, and everything else that techwebs do. Can you guess which disk does what in this screenshot?  ## Changelog 🆑 balance: You can no longer print Technology data disks. You can still print Tech disks, which hold techweb information on it, just not the one that holds up to 5 nodes. balance: ^ Because of this, there's no way to download nodes from an RD console and upload them to an Autolathe to bypass departmental restrictions, you have to go through a Techfab/Circuit imprinter for your needs. balance: Ones that are found cannot have anything uploaded/deleted off of them either, you can only upload them to the Web. code: Every individual Bepis disk no longer create an entire techweb /🆑
520 lines
18 KiB
Plaintext
520 lines
18 KiB
Plaintext
/**
|
|
* # Techweb
|
|
*
|
|
* A datum representing a research techweb
|
|
*
|
|
* Techweb datums are meant to store unlocked research, being able to be stored
|
|
* on research consoles, servers, and disks. They are NOT global.
|
|
*/
|
|
/datum/techweb
|
|
///The id/name of the whole Techweb viewable to players.
|
|
var/id = "Generic"
|
|
/// Organization name, used for display
|
|
var/organization = "Third-Party"
|
|
|
|
/// Already unlocked and all designs are now available. Assoc list, id = TRUE
|
|
var/list/researched_nodes = list()
|
|
/// Visible nodes, doesn't mean it can be researched. Assoc list, id = TRUE
|
|
var/list/visible_nodes = list()
|
|
/// Nodes that can immediately be researched, all reqs met. assoc list, id = TRUE
|
|
var/list/available_nodes = list()
|
|
/// Designs that are available for use. Assoc list, id = TRUE
|
|
var/list/researched_designs = list()
|
|
/// Custom inserted designs like from disks that should survive recalculation.
|
|
var/list/custom_designs = list()
|
|
/// Already boosted nodes that can't be boosted again. node id = path of boost object.
|
|
var/list/boosted_nodes = list()
|
|
/// Hidden nodes. id = TRUE. Used for unhiding nodes when requirements are met by removing the entry of the node.
|
|
var/list/hidden_nodes = list()
|
|
/// Items already deconstructed for a generic point boost, path = list(point_type = points)
|
|
var/list/deconstructed_items = list()
|
|
/// Available research points, type = number
|
|
var/list/research_points = list()
|
|
/// Game logs of research nodes, "node_name" "node_cost" "node_researcher" "node_research_location"
|
|
var/list/research_logs = list()
|
|
/// Current per-second production, used for display only.
|
|
var/list/last_bitcoins = list()
|
|
/// Mutations discovered by genetics, this way they are shared and cant be destroyed by destroying a single console
|
|
var/list/discovered_mutations = list()
|
|
/// Assoc list, id = number, 1 is available, 2 is all reqs are 1, so on
|
|
var/list/tiers = list()
|
|
/// This is a list of all incomplete experiment datums that are accessible for scientists to complete
|
|
var/list/datum/experiment/available_experiments = list()
|
|
/// A list of all experiment datums that have been complete
|
|
var/list/datum/experiment/completed_experiments = list()
|
|
/// Assoc list of all experiment datums that have been skipped, to tech point reward for completing them -
|
|
/// That is, upon researching a node without completing its associated discounts, their experiments go here.
|
|
/// Completing these experiments will have a refund.
|
|
var/list/datum/experiment/skipped_experiment_types = list()
|
|
|
|
/// If science researches something without completing its discount experiments,
|
|
/// they have the option to complete them later for a refund
|
|
/// This ratio determines how much of the original discount is refunded
|
|
var/skipped_experiment_refund_ratio = 0.66
|
|
|
|
///All RD consoles connected to this individual techweb.
|
|
var/list/obj/machinery/computer/rdconsole/consoles_accessing = list()
|
|
///All research servers connected to this individual techweb.
|
|
var/list/obj/machinery/rnd/server/techweb_servers = list()
|
|
|
|
///Boolean on whether the techweb should generate research points overtime.
|
|
var/should_generate_points = FALSE
|
|
///A multiplier applied to all research gain, cut in half if the Master server was sabotaged.
|
|
var/income_modifier = 1
|
|
///The amount of research points generated the techweb generated the latest time it generated.
|
|
var/last_income
|
|
|
|
/**
|
|
* Assoc list of relationships with various partners
|
|
* scientific_cooperation[partner_typepath] = relationship
|
|
*/
|
|
var/list/scientific_cooperation
|
|
/**
|
|
* Assoc list of papers already published by the crew.
|
|
* published_papers[experiment_typepath][tier] = paper
|
|
* Filled with nulls on init, populated only on publication.
|
|
*/
|
|
var/list/published_papers
|
|
|
|
/datum/techweb/New()
|
|
SSresearch.techwebs += src
|
|
for(var/i in SSresearch.techweb_nodes_starting)
|
|
var/datum/techweb_node/DN = SSresearch.techweb_node_by_id(i)
|
|
research_node(DN, TRUE, FALSE, FALSE)
|
|
hidden_nodes = SSresearch.techweb_nodes_hidden.Copy()
|
|
initialize_published_papers()
|
|
return ..()
|
|
|
|
/datum/techweb/Destroy()
|
|
researched_nodes = null
|
|
researched_designs = null
|
|
available_nodes = null
|
|
visible_nodes = null
|
|
custom_designs = null
|
|
SSresearch.techwebs -= src
|
|
return ..()
|
|
|
|
/datum/techweb/proc/recalculate_nodes(recalculate_designs = FALSE, wipe_custom_designs = FALSE)
|
|
var/list/datum/techweb_node/processing = list()
|
|
for(var/id in researched_nodes)
|
|
processing[id] = TRUE
|
|
for(var/id in visible_nodes)
|
|
processing[id] = TRUE
|
|
for(var/id in available_nodes)
|
|
processing[id] = TRUE
|
|
if(recalculate_designs)
|
|
researched_designs = custom_designs.Copy()
|
|
if(wipe_custom_designs)
|
|
custom_designs = list()
|
|
for(var/id in processing)
|
|
update_node_status(SSresearch.techweb_node_by_id(id))
|
|
CHECK_TICK
|
|
|
|
/datum/techweb/proc/add_point_list(list/pointlist)
|
|
for(var/i in pointlist)
|
|
if(SSresearch.point_types[i] && pointlist[i] > 0)
|
|
research_points[i] += pointlist[i]
|
|
|
|
/datum/techweb/proc/add_points_all(amount)
|
|
var/list/l = SSresearch.point_types.Copy()
|
|
for(var/i in l)
|
|
l[i] = amount
|
|
add_point_list(l)
|
|
|
|
/datum/techweb/proc/remove_point_list(list/pointlist)
|
|
for(var/i in pointlist)
|
|
if(SSresearch.point_types[i] && pointlist[i] > 0)
|
|
research_points[i] = max(0, research_points[i] - pointlist[i])
|
|
|
|
/datum/techweb/proc/remove_points_all(amount)
|
|
var/list/l = SSresearch.point_types.Copy()
|
|
for(var/i in l)
|
|
l[i] = amount
|
|
remove_point_list(l)
|
|
|
|
/datum/techweb/proc/modify_point_list(list/pointlist)
|
|
for(var/i in pointlist)
|
|
if(SSresearch.point_types[i] && pointlist[i] != 0)
|
|
research_points[i] = max(0, research_points[i] + pointlist[i])
|
|
|
|
/datum/techweb/proc/modify_points_all(amount)
|
|
var/list/l = SSresearch.point_types.Copy()
|
|
for(var/i in l)
|
|
l[i] = amount
|
|
modify_point_list(l)
|
|
|
|
/datum/techweb/proc/copy_research_to(datum/techweb/receiver) //Adds any missing research to theirs.
|
|
for(var/i in receiver.hidden_nodes)
|
|
CHECK_TICK
|
|
if(available_nodes[i] || researched_nodes[i] || visible_nodes[i])
|
|
receiver.hidden_nodes -= i //We can see it so let them see it too.
|
|
for(var/i in researched_nodes)
|
|
CHECK_TICK
|
|
receiver.research_node_id(i, TRUE, FALSE, FALSE)
|
|
for(var/i in researched_designs)
|
|
CHECK_TICK
|
|
receiver.add_design_by_id(i)
|
|
receiver.recalculate_nodes()
|
|
|
|
/datum/techweb/proc/copy()
|
|
var/datum/techweb/returned = new()
|
|
returned.researched_nodes = researched_nodes.Copy()
|
|
returned.visible_nodes = visible_nodes.Copy()
|
|
returned.available_nodes = available_nodes.Copy()
|
|
returned.researched_designs = researched_designs.Copy()
|
|
returned.hidden_nodes = hidden_nodes.Copy()
|
|
return returned
|
|
|
|
/datum/techweb/proc/get_visible_nodes() //The way this is set up is shit but whatever.
|
|
return visible_nodes - hidden_nodes
|
|
|
|
/datum/techweb/proc/get_available_nodes()
|
|
return available_nodes - hidden_nodes
|
|
|
|
/datum/techweb/proc/get_researched_nodes()
|
|
return researched_nodes - hidden_nodes
|
|
|
|
/datum/techweb/proc/add_point_type(type, amount)
|
|
if(!SSresearch.point_types[type] || (amount <= 0))
|
|
return FALSE
|
|
research_points[type] += amount
|
|
return TRUE
|
|
|
|
/datum/techweb/proc/modify_point_type(type, amount)
|
|
if(!SSresearch.point_types[type])
|
|
return FALSE
|
|
research_points[type] = max(0, research_points[type] + amount)
|
|
return TRUE
|
|
|
|
/datum/techweb/proc/remove_point_type(type, amount)
|
|
if(!SSresearch.point_types[type] || (amount <= 0))
|
|
return FALSE
|
|
research_points[type] = max(0, research_points[type] - amount)
|
|
return TRUE
|
|
|
|
/**
|
|
* add_design_by_id
|
|
* The main way to add add designs to techweb
|
|
* Uses the techweb node's ID
|
|
* Args:
|
|
* id - the ID of the techweb node to research
|
|
* custom - Boolean on whether the node should also be added to custom_designs
|
|
* add_to - A custom list to add the node to, overwriting research_designs.
|
|
*/
|
|
/datum/techweb/proc/add_design_by_id(id, custom = FALSE, list/add_to)
|
|
return add_design(SSresearch.techweb_design_by_id(id), custom, add_to)
|
|
|
|
/datum/techweb/proc/add_design(datum/design/design, custom = FALSE, list/add_to)
|
|
if(!istype(design))
|
|
return FALSE
|
|
SEND_SIGNAL(src, COMSIG_TECHWEB_ADD_DESIGN, design, custom)
|
|
if(custom)
|
|
custom_designs[design.id] = TRUE
|
|
|
|
if(add_to)
|
|
add_to[design.id] = TRUE
|
|
else
|
|
researched_designs[design.id] = TRUE
|
|
|
|
hidden_nodes -= design.id
|
|
|
|
return TRUE
|
|
|
|
/datum/techweb/proc/remove_design_by_id(id, custom = FALSE)
|
|
return remove_design(SSresearch.techweb_design_by_id(id), custom)
|
|
|
|
/datum/techweb/proc/remove_design(datum/design/design, custom = FALSE)
|
|
if(!istype(design))
|
|
return FALSE
|
|
if(custom_designs[design.id] && !custom)
|
|
return FALSE
|
|
SEND_SIGNAL(src, COMSIG_TECHWEB_REMOVE_DESIGN, design, custom)
|
|
custom_designs -= design.id
|
|
researched_designs -= design.id
|
|
return TRUE
|
|
|
|
/datum/techweb/proc/get_point_total(list/pointlist)
|
|
for(var/i in pointlist)
|
|
. += pointlist[i]
|
|
|
|
/datum/techweb/proc/can_afford(list/pointlist)
|
|
for(var/i in pointlist)
|
|
if(research_points[i] < pointlist[i])
|
|
return FALSE
|
|
return TRUE
|
|
|
|
/**
|
|
* Checks if all experiments have been completed for a given node on this techweb
|
|
*
|
|
* Arguments:
|
|
* * node - the node to check
|
|
*/
|
|
/datum/techweb/proc/have_experiments_for_node(datum/techweb_node/node)
|
|
. = TRUE
|
|
for (var/experiment_type in node.required_experiments)
|
|
if (!completed_experiments[experiment_type])
|
|
return FALSE
|
|
|
|
/**
|
|
* Checks if a node can be unlocked on this techweb, having the required points and experiments
|
|
*
|
|
* Arguments:
|
|
* * node - the node to check
|
|
*/
|
|
/datum/techweb/proc/can_unlock_node(datum/techweb_node/node)
|
|
return can_afford(node.get_price(src)) && have_experiments_for_node(node)
|
|
|
|
/**
|
|
* Adds an experiment to this techweb by its type, ensures that no duplicates are added.
|
|
*
|
|
* Arguments:
|
|
* * experiment_type - the type of the experiment to add
|
|
*/
|
|
/datum/techweb/proc/add_experiment(experiment_type)
|
|
. = TRUE
|
|
// check active experiments for experiment of this type
|
|
for (var/available_experiment in available_experiments)
|
|
var/datum/experiment/experiment = available_experiment
|
|
if (experiment.type == experiment_type)
|
|
return FALSE
|
|
// check completed experiments for experiments of this type
|
|
for (var/completed_experiment in completed_experiments)
|
|
var/datum/experiment/experiment = completed_experiment
|
|
if (experiment == experiment_type)
|
|
return FALSE
|
|
available_experiments += new experiment_type()
|
|
|
|
/**
|
|
* Adds a list of experiments to this techweb by their types, ensures that no duplicates are added.
|
|
*
|
|
* Arguments:
|
|
* * experiment_list - the list of types of experiments to add
|
|
*/
|
|
/datum/techweb/proc/add_experiments(list/experiment_list)
|
|
. = TRUE
|
|
for (var/datum/experiment/experiment as anything in experiment_list)
|
|
. = . && add_experiment(experiment)
|
|
|
|
/**
|
|
* Notifies the techweb that an experiment has been completed, updating internal state of the techweb to reflect this.
|
|
*
|
|
* Arguments:
|
|
* * completed_experiment - the experiment which was completed
|
|
*/
|
|
/datum/techweb/proc/complete_experiment(datum/experiment/completed_experiment)
|
|
available_experiments -= completed_experiment
|
|
completed_experiments[completed_experiment.type] = completed_experiment
|
|
|
|
var/result_text = "[completed_experiment] has been completed"
|
|
var/refund = skipped_experiment_types[completed_experiment.type] || 0
|
|
if(refund > 0)
|
|
add_point_list(list(TECHWEB_POINT_TYPE_GENERIC = refund))
|
|
result_text += ", refunding [refund] points."
|
|
// Nothing more to gain here, but we keep it in the list to prevent double dipping
|
|
skipped_experiment_types[completed_experiment.type] = -1
|
|
else
|
|
result_text += "!"
|
|
|
|
log_research("[completed_experiment.name] ([completed_experiment.type]) has been completed on techweb [id]/[organization][refund ? ", refunding [refund] points" : ""].")
|
|
return result_text
|
|
|
|
/datum/techweb/proc/printout_points()
|
|
return techweb_point_display_generic(research_points)
|
|
|
|
/datum/techweb/proc/research_node_id(id, force, auto_update_points, get_that_dosh_id)
|
|
return research_node(SSresearch.techweb_node_by_id(id), force, auto_update_points, get_that_dosh_id)
|
|
|
|
/datum/techweb/proc/research_node(datum/techweb_node/node, force = FALSE, auto_adjust_cost = TRUE, get_that_dosh = TRUE)
|
|
if(!istype(node))
|
|
return FALSE
|
|
update_node_status(node)
|
|
if(!force)
|
|
if(!available_nodes[node.id] || (auto_adjust_cost && (!can_afford(node.get_price(src)))) || !have_experiments_for_node(node))
|
|
return FALSE
|
|
var/log_message = "[id]/[organization] researched node [node.id]"
|
|
if(auto_adjust_cost)
|
|
var/list/node_cost = node.get_price(src)
|
|
remove_point_list(node_cost)
|
|
log_message += " at the cost of [json_encode(node_cost)]"
|
|
|
|
//Add to our researched list
|
|
researched_nodes[node.id] = TRUE
|
|
|
|
// Track any experiments we skipped relating to this
|
|
for(var/missed_experiment in node.discount_experiments)
|
|
if(completed_experiments[missed_experiment] || skipped_experiment_types[missed_experiment])
|
|
continue
|
|
skipped_experiment_types[missed_experiment] = node.discount_experiments[missed_experiment] * skipped_experiment_refund_ratio
|
|
|
|
// Gain the experiments from the new node
|
|
for(var/id in node.unlock_ids)
|
|
visible_nodes[id] = TRUE
|
|
var/datum/techweb_node/unlocked_node = SSresearch.techweb_node_by_id(id)
|
|
if (unlocked_node.required_experiments.len > 0)
|
|
add_experiments(unlocked_node.required_experiments)
|
|
if (unlocked_node.discount_experiments.len > 0)
|
|
add_experiments(unlocked_node.discount_experiments)
|
|
update_node_status(unlocked_node)
|
|
|
|
// Unlock what the research actually unlocks
|
|
for(var/id in node.design_ids)
|
|
add_design_by_id(id)
|
|
update_node_status(node)
|
|
if(get_that_dosh)
|
|
var/datum/bank_account/science_department_bank_account = SSeconomy.get_dep_account(ACCOUNT_SCI)
|
|
science_department_bank_account?.adjust_money(SSeconomy.techweb_bounty)
|
|
log_message += ", gaining [SSeconomy.techweb_bounty] to [science_department_bank_account] for it."
|
|
|
|
// Avoid logging the same 300+ lines at the beginning of every round
|
|
if (MC_RUNNING())
|
|
log_research(log_message)
|
|
|
|
return TRUE
|
|
|
|
/datum/techweb/proc/unresearch_node_id(id)
|
|
return unresearch_node(SSresearch.techweb_node_by_id(id))
|
|
|
|
/datum/techweb/proc/unresearch_node(datum/techweb_node/node)
|
|
if(!istype(node))
|
|
return FALSE
|
|
researched_nodes -= node.id
|
|
recalculate_nodes(TRUE) //Fully rebuild the tree.
|
|
|
|
/// Boosts a techweb node.
|
|
/datum/techweb/proc/boost_techweb_node(datum/techweb_node/node, list/pointlist)
|
|
if(!istype(node))
|
|
return FALSE
|
|
LAZYINITLIST(boosted_nodes[node.id])
|
|
for(var/point_type in pointlist)
|
|
boosted_nodes[node.id][point_type] = max(boosted_nodes[node.id][point_type], pointlist[point_type])
|
|
if(node.autounlock_by_boost)
|
|
hidden_nodes -= node.id
|
|
update_node_status(node)
|
|
return TRUE
|
|
|
|
/// Boosts a techweb node by using items.
|
|
/datum/techweb/proc/boost_with_item(datum/techweb_node/node, itempath)
|
|
if(!istype(node) || !ispath(itempath))
|
|
return FALSE
|
|
var/list/boost_amount = node.boost_item_paths[itempath]
|
|
boost_techweb_node(node, boost_amount)
|
|
return TRUE
|
|
|
|
/datum/techweb/proc/update_tiers(datum/techweb_node/base)
|
|
var/list/current = list(base)
|
|
while (current.len)
|
|
var/list/next = list()
|
|
for (var/node_ in current)
|
|
var/datum/techweb_node/node = node_
|
|
var/tier = 0
|
|
if (!researched_nodes[node.id]) // researched is tier 0
|
|
for (var/id in node.prereq_ids)
|
|
var/prereq_tier = tiers[id]
|
|
tier = max(tier, prereq_tier + 1)
|
|
|
|
if (tier != tiers[node.id])
|
|
tiers[node.id] = tier
|
|
for (var/id in node.unlock_ids)
|
|
next += SSresearch.techweb_node_by_id(id)
|
|
current = next
|
|
|
|
/datum/techweb/proc/update_node_status(datum/techweb_node/node)
|
|
var/researched = FALSE
|
|
var/available = FALSE
|
|
var/visible = FALSE
|
|
if(researched_nodes[node.id])
|
|
researched = TRUE
|
|
var/needed = node.prereq_ids.len
|
|
for(var/id in node.prereq_ids)
|
|
if(researched_nodes[id])
|
|
visible = TRUE
|
|
needed--
|
|
if(!needed)
|
|
available = TRUE
|
|
researched_nodes -= node.id
|
|
available_nodes -= node.id
|
|
visible_nodes -= node.id
|
|
if(hidden_nodes[node.id]) //Hidden.
|
|
return
|
|
if(researched)
|
|
researched_nodes[node.id] = TRUE
|
|
for(var/id in node.design_ids)
|
|
add_design(SSresearch.techweb_design_by_id(id))
|
|
else
|
|
if(available)
|
|
available_nodes[node.id] = TRUE
|
|
else
|
|
if(visible)
|
|
visible_nodes[node.id] = TRUE
|
|
update_tiers(node)
|
|
|
|
//Laggy procs to do specific checks, just in case. Don't use them if you can just use the vars that already store all this!
|
|
/datum/techweb/proc/designHasReqs(datum/design/D)
|
|
for(var/i in researched_nodes)
|
|
var/datum/techweb_node/N = SSresearch.techweb_node_by_id(i)
|
|
if(N.design_ids[D.id])
|
|
return TRUE
|
|
return FALSE
|
|
|
|
/datum/techweb/proc/isDesignResearched(datum/design/D)
|
|
return isDesignResearchedID(D.id)
|
|
|
|
/datum/techweb/proc/isDesignResearchedID(id)
|
|
return researched_designs[id]? SSresearch.techweb_design_by_id(id) : FALSE
|
|
|
|
/datum/techweb/proc/isNodeResearched(datum/techweb_node/N)
|
|
return isNodeResearchedID(N.id)
|
|
|
|
/datum/techweb/proc/isNodeResearchedID(id)
|
|
return researched_nodes[id]? SSresearch.techweb_node_by_id(id) : FALSE
|
|
|
|
/datum/techweb/proc/isNodeVisible(datum/techweb_node/N)
|
|
return isNodeResearchedID(N.id)
|
|
|
|
/datum/techweb/proc/isNodeVisibleID(id)
|
|
return visible_nodes[id]? SSresearch.techweb_node_by_id(id) : FALSE
|
|
|
|
/datum/techweb/proc/isNodeAvailable(datum/techweb_node/N)
|
|
return isNodeAvailableID(N.id)
|
|
|
|
/datum/techweb/proc/isNodeAvailableID(id)
|
|
return available_nodes[id]? SSresearch.techweb_node_by_id(id) : FALSE
|
|
|
|
/// Fill published_papers with nulls.
|
|
/datum/techweb/proc/initialize_published_papers()
|
|
published_papers = list()
|
|
scientific_cooperation = list()
|
|
for (var/datum/experiment/ordnance/ordnance_experiment as anything in SSresearch.ordnance_experiments)
|
|
var/max_tier = min(length(ordnance_experiment.gain), length(ordnance_experiment.target_amount))
|
|
var/list/tier_list[max_tier]
|
|
published_papers[ordnance_experiment.type] = tier_list
|
|
for (var/datum/scientific_partner/partner as anything in SSresearch.scientific_partners)
|
|
scientific_cooperation[partner.type] = 0
|
|
|
|
/// Publish the paper into our techweb. Cancel if we are not allowed to.
|
|
/datum/techweb/proc/add_scientific_paper(datum/scientific_paper/paper_to_add)
|
|
if(!paper_to_add.allowed_to_publish(src))
|
|
return FALSE
|
|
paper_to_add.publish_paper(src)
|
|
|
|
// If we haven't published a paper in the same topic ...
|
|
if(locate(paper_to_add.experiment_path) in published_papers[paper_to_add.experiment_path])
|
|
return TRUE
|
|
// Quickly add and complete it.
|
|
// PS: It's also possible to use add_experiment() together with a list/available_experiments check
|
|
// to determine if we need to run all this, but this pretty much does the same while only needing one evaluation.
|
|
|
|
add_experiment(paper_to_add.experiment_path)
|
|
|
|
for (var/datum/experiment/experiment as anything in available_experiments)
|
|
if(experiment.type != paper_to_add.experiment_path)
|
|
continue
|
|
|
|
experiment.completed = TRUE
|
|
var/announcetext = complete_experiment(experiment)
|
|
if(length(GLOB.experiment_handlers))
|
|
var/datum/component/experiment_handler/handler = GLOB.experiment_handlers[1]
|
|
handler.announce_message_to_all(announcetext)
|
|
|
|
return TRUE
|